Articles
- Page Path
- HOME > Kosin Med J > Volume 37(3); 2022 > Article
-
Original article
Identification of the transcriptome profile of Miamiensis avidus after mebendazole treatment -
Hyunsu Kim*, A-Reum Lee*, Kyung-Yoon Jeon, Eun-Ji Ko
 , Hee-Jae Cha**
, Hee-Jae Cha** , Mee Sun Ock**
, Mee Sun Ock**
-
Kosin Medical Journal 2022;37(3):203-212.
DOI: https://doi.org/10.7180/kmj.22.003
Published online: May 16, 2022
Department of Parasitology and Genetics, Institute for Medical Science, Kosin University College of Medicine, Busan, Korea
- Corresponding Author: Hee-Jae Cha, PhD Department of Parasitology and Genetics, Kosin University College of Medicine, 262 Gamcheon-ro, Seo-gu, Busan 49267, Korea Tel: +82-51-990-6428 Fax: +82-51-990-3081 E-mail: hcha@kosin.ac.kr
- Corresponding Author: Mee Sun Ock, PhD Department of Parasitology and Genetics, Kosin University College of Medicine, 262 Gamcheon-ro, Seo-gu, Busan 49267, Korea Tel: +82-51-990-6424 Fax: +82-51-990-3081 E-mail: sunnyock@kosin.ac.kr
- *These authors contributed equally to this work as first authors.
**These authors contributed equally to this work as corresponding authors.
Copyright © 2022 Kosin University College of Medicine.
This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 1,785 Views
- 37 Download
Abstract
-
Background
- The scuticociliate Miamiensis avidus is a major pathogenic agent that causes significant economic losses in the flounder aquaculture industry. Many different types of drugs are being tested to control this disease, including mebendazole, which is a broad-spectrum antiprotozoal agent. The purpose of this study was to determine whether mebendazole worked in vitro against M. avidus and to explore its mechanism of action.
-
Methods
- Transcriptome and gene ontology analyses were conducted to investigate the specifically expressed gene profile. We confirmed the cytotoxic effect of mebendazole against M. avidus when it was applied intermittently for a total of three times. We also identified differentially expressed genes using transcriptome analysis.
-
Results
- Most of the upregulated genes were membrane transport-related genes, including Na+/K+-ATPase. Most of the downregulated genes were categorized into three groups: tubulin-related, metabolism-related, and transport-related genes. The expression levels of glucose uptake-related genes decreased due to the inhibition of tubulin polymerization, but this was not statistically significant.
-
Conclusions
- Our results demonstrate that intermittent treatment with mebendazole has a significant cytotoxic effect on M. avidus. Furthermore, mebendazole induces downregulation of the tubulin-alpha chain and metabolism-related genes. It is presumed that this leads to a glucose shortage and the death of M. avidus. Transcriptome analysis will provide useful clues for further studies on mebendazole applications for scutica control.
- Scuticociliatosis, a parasitic disease caused by invasive ciliates (class: Scuticociliatida), has the largest detrimental impact on the fish industry. In South Korea, scuticociliatosis was first identified in an olive flounder farm on Jeju Island in the 1990s, and is now known to cause serious economic damage to olive flounders in farms nationwide every year [1,2]. Since the first report of scuticociliates as parasites of seahorses, they have been reported to infect various species of marine animals, causing serious damage [3]. Among closely related pathogenic ciliate species, Miamiensis avidus was identified as the dominant species with the strongest pathogenicity in olive flounders and other cultured fish [4].
- Although several drug candidates are currently in trials, the Korean government had granted item permissions for the use of formalin (37% formaldehyde) in 2006 and hydrogen peroxide (35% hydrogen peroxide) in 2015 for olive flounder farms. The two agents have proven to be effective drugs for the treatment of scuticociliate infection in multiple studies, and they were subsequently commercialized [5]. However, there is a need for drugs that can effectively treat scutica without adversely affecting the host fish or marine environment.
- Benzimidazole derivatives, such as mebendazole and albendazole, have been used as anthelmintics worldwide, in both human and veterinary medicine, for the treatment of various helminth infections [6,7]. Recently, mebendazole and albendazole have been repositioned as promising anti-cancer agents [8]. Among these benzimidazole derivatives, mebendazole has a similar therapeutic effect as albendazole, but has been found to induce only milder oxidative stress than albendazole in the hosts, and it was presumed to be the drug of choice for the treatment of parasitic protozoa and helminths [9]. Mebendazole has also been reported to have antiparasitic effects on monogeneans and scuticociliates that are parasitic on the gills or tissues of various aquatic species [10,11].
- Therefore, we evaluated the efficacy of mebendazole against M. avidus in vitro and investigated its gene expression profile during the killing process of mebendazole treatment via transcriptome analysis.
Introduction
- 1. Parasite strains and cultivation
- The ciliates used in this study were obtained from Pukyong National University (Busan, Korea), which were identified to be M. avidus using species-specific oligonucleotide primers reported [12]. M. avidus was inoculated into a culture medium with 2% peptone (BD Biosciences, Franklin Lakes, NJ, USA), 1% yeast extract (BD Biosciences), 0.5% sodium chloride (BD Biosciences), 10% fetal bovine serum (Biowest, Riverside, MO, USA), and 1% penicillin-streptomycin (Gibco, Carlsbad, CA, USA) for 3 to 5 days at 22°C.
- 2. In vitro anti-M. avidus activity of mebendazole
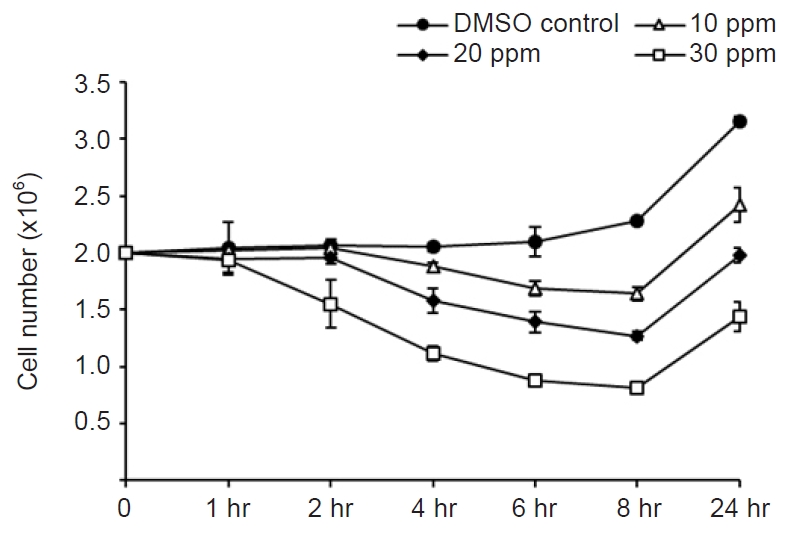
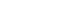
- Mebendazole (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) at a concentration of 3,000 ppm. For the antiparasitic activity test, 5 mL (2×106) of M. avidus was dispensed into a 25 cm2 flask, and 10, 20, and 30 ppm concentrations of mebendazole were added. Control cells were treated with DMSO only. Then, the number of M. avidus was counted after 1, 2, 4, 6, and 8 hours. The amount of DMSO did not exceed 1% of the total volume. Our preliminary test revealed that DMSO (<1%) did not show any harmful effect against M. avidus (data not shown).
- Among the tested concentrations, 30 ppm showed the highest cytotoxic effect against M. avidus. After 8 hours of treatment with 30 ppm of mebendazole, 50%–60% of M. avidus species were killed, but the number of M. avidus cells began to increase after 8 hours. To increase the cytotoxicity to 100%, M. avidus was treated with 30 ppm mebendazole at 4 and 8 hours. We counted the number of live M. avidus (actively moving with cilia) after 24 hours.
- 3. Library preparation and strand-specific RNA-sequencing
- When we treated M. avidus with 30 ppm of mebendazole for 8 hours, the mortality rate was found to be 50%–60%. Therefore, we used this group as the treated group.
- First, 10–20 mL of scutica culture was pelleted by centrifugation at 3,500 ×g for 10 minutes at room temperature. Then, 2–4 mL of RNAprotect (Qiagen, Hilden, Germany) was added to the pellet and mixed by vortexing. The pellet was re-precipitated by centrifugation at 3,500 ×g for 10 minutes at room temperature, and the supernatant was discarded. The standard TRIzol (Life Technologies, Carlsbad, CA, USA) protocol was then applied to extract the total RNA from cell pellets. To ensure that the DNA was completely removed, DNase digestion was performed, and the total RNA samples were further purified using acidic phenol–chloroform. The sequencing libraries were prepared using an RNA-seq Library Preparation kit (Epicentre Biotechnologies, Madison, WI, USA) with rRNA-depleted samples, and all of the libraries were sequenced by Illumina HiSeq 2000 following the strand-specific sequencing protocol for 100 cycles.
- 4. Reads processing and expression calculation
- The first six bases of reads and adaptors were removed using in-house-developed pipelines. The transcripts were assembled using the default parameters. All transcripts were annotated with coverage and identity greater than or equal to 0.8. Overlapped annotations on transcripts were further combined if they overlapped with each other by at least 70% of their lengths. Based on the gene annotations of the transcripts, the reads per kilobase per million mapped reads (RPKM) values were calculated to determine the gene expression levels.
- 5. Differential expression determination
- Differentially expressed genes (DEGs) between the mebendazole-treated and control groups were determined using the following set. To be considered as an upregulation under drug treatment, the normalized expression value of the gene in the treated sample at mid-log phase must be larger than or equal to 50 RPKM. To be considered as a downregulation, the normalized expression value of the gene in the control at mid-log phase must be greater than or equal to 50 RPKM. For upregulation under drug treatment, the normalized expression value of the gene in the treated group at mid-log phase must be at least 2-fold larger than that in the control; for downregulation under mebendazole treatment, the normalized expression value of the gene in the control group at mid-log phase must be at least 2-fold larger than that in the treated group.
- 6. GO and KEGG enrichment analyses of DEGs
- Gene ontology (GO) enrichment analysis of DEGs was performed using the GO seq R package, in which gene length bias was corrected. GO terms with corrected p-values <0.05, were considered to be significantly enriched in DEGs. KOBAS 2.0 was used to test the statistical enrichment of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
Methods
- 1. Anti-M. avidus activity of Mebendazole in vitro
- The concentration of 30 ppm mebendazole showed the highest killing effect against M. avidus after 8 hours of incubation (Fig. 1). However, the number of M. avidus began to increase after 8 hours. After the application of mebendazole at 4 and 8 hours after the first treatment, the killing effect of more than 99.9% was confirmed after 24 hours (Fig. 2). This confirmed that mebendazole was effective in killing M. avidus in vitro.
- 2. RNA sequencing and aligning to the reference genome
- To investigate the gene expression profile associated with the cytotoxic effect of mebendazole against M. avidus, transcriptome analysis was carried out. The two cDNA libraries (control and mebendazole-treated groups) were sequenced on an Illumina HiSeq 4000 platform. A total of 98,172,410 and 53,616,548 raw reads were generated from the control and mebendazole-treated groups’ databases, respectively (Tables 1, 2). After removing the low-quality reads, 95,425,100 and 52,123,958 clean readings were obtained, mapping 97.2% and 97.2%, respectively.
- 3. Gene functional annotation
- The presumptive annotation of these transcripts was performed using BLASTX. The putative functions of 11,253 (64.8%) sequences of 17,346 unigene sequences in the control group and 12,167 (53.8%) sequences among 22,631 unigene sequences in the mebendazole-treated group were confirmed (Tables 1, 2).
- 4. Specific gene expression after mebendazole treatment
- There were 204 DEGs with significant differences among 10,621 DEGs. Among them, 48 DEGs were upregulated, while 156 DEGs were downregulated. The most abundantly expressed genes in the control (untreated) and treated groups were the 40S and 60S ribosomal protein genes (Table 3). Transcripts related to the channel and transport proteins, such as Na+/K+ ATPase alpha subunit, aquaporin, polyol transporter 2, transmembrane amino acid transporter protein, and granule lattice protein 3 precursor, were upregulated (Table 4). Table 4 also shows the top 20 ranked downregulated genes. Among them, three categories of gene groups known to be associated with the action mechanism of mebendazole were identified (Table 5). In addition, the expression of pyruvate carboxylase related to glucose metabolism was downregulated. Catalase and peroxidase, which are important enzymes that protect the cells from oxidative damage by reactive oxygen species, were also downregulated (Table 5). Glucose transport and immune response-related genes were also identified (Table 6). However, most of them were not significant statistically.
- 5. GO enrichment analysis of DEGs
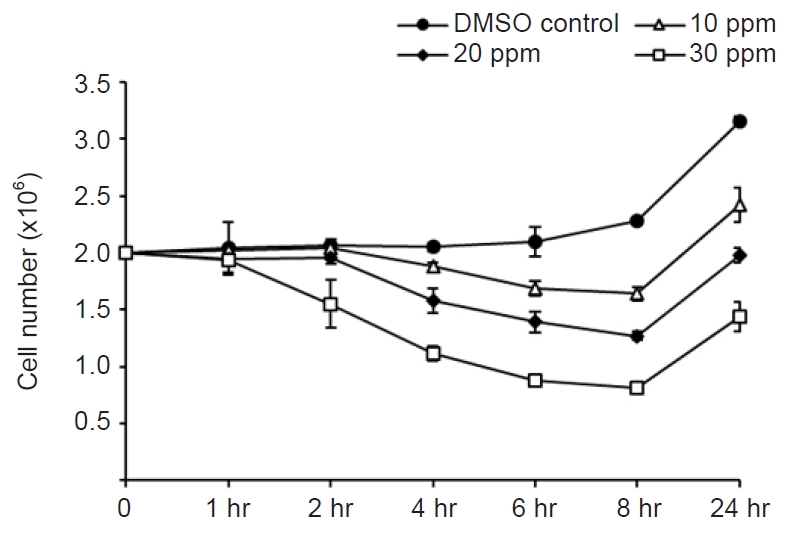
- GO enrichment is commonly used to explain the biological roles of genes and their products. All DEGs were mapped to GO database terminology and compared to the overall transcriptome background to determine the functionality of the DEGs. All DEGs were categorized into three major functional categories: biological processes (565 unigenes), cellular components (769 unigenes), and molecular functions (210 unigenes) (Fig. 3).
- The cellular process (124 unigenes), metabolic process (116 unigenes), and single-organism process (93 unigenes) represented the majority category of biological processes. The majority of cellular components were composed of cells (144 unigenes), cell parts (144 unigenes), membranes (98 unigenes), and organelles (127 unigenes).
- Binding (100 unigenes) and catalytic activity (76 unigenes) accounted for the largest portion of the molecular function categories.
Results
- Here, we report the profile of DEGs and GO analysis after mebendazole treatment of M. avidus. Our data clearly indicate that mebendazole has significant deleterious effects on M. avidus.
- When M. avidus ciliate was treated with mebendazole (30 ppm) for 4 hours, more than 50% of scutica was killed and the survival rate dropped to less than 40% at 8 hours. However, the number of live cells started to increase thereafter. In order to increase the killing effect of mebendazole, the drug was treated twice at 4 and 8 hours after the first mebendazole treatment. After 24 hours, most of the scutica cells were killed. These results suggest that the appropriate intermittent administration of mebendazole can be effective for the control of scutica. However, the low efficacy of mebendazole in seawater seems to be one of the most important challenges in its application [13].
- The mechanism underlying the antiparasitic action of mebendazole is known to inhibit tubulin polymerization and the formation of microtubules. Glucose transporter (GLUT)-2 is also blocked by mebendazole, which prevents glucose uptake by parasites in the intestines [14,15]. In addition, mebendazole has been reported to activate the mitogen-activated protein kinase (MEK)-extracellular signal‐regulated kinase (ERK) pathway in tubulin-activating drugs [16]. We also confirmed the changes in the gene expression patterns of three key categories: membrane transport-related, tubulin-related, and metabolic processes. The characteristic changes in these three categories are in good agreement with the previously known the action mechanism of mebendazole.
- Among the upregulated genes, expression of membrane channel and transport-related genes, such as Na+/K+ ATPase alpha subunit, apolipoprotein, aquaporin, polyol transporter 2, and transmembrane amino acid transporter protein, were confirmed. Granule lattice protein precursor transcripts are highly upregulated, and these genes are known to be involved in protein sorting and solubility [17,18]. Mebendazole is thought to primarily affect the membrane. Membrane damage and permeability changes caused by mebendazole treatment have been confirmed by antifungal activity screening in previous reports [19,20]. It seems that the expression of related genes was increased to restore the damaged membranes and altered permeability.
- The downregulated genes were categorized into three main groups: microtubule-related, metabolism-related and transport-related genes. Genes belonging to the microtubule-related group were microtubule constituent genes, tubulin-binding protein transcript, lactoylglutathione lyase, and dynein. Lactoylglutathione lyase is known to be associated with microtubule assembly, and dynein is a protein family responsible for the movement of cilia and flagella by moving along microtubules [21,22]. Our results on microtubule-related genes were consistent with those of previous reports.
- Expression levels of genes belonging to the energy metabolism-related group, such as serine hydroxymethyltransferase 1, pyruvate carboxylase, phosphoglycerate kinase 1, enolase, and sodium/calcium exchanger protein, were significantly decreased. It is well known that inhibition of tubulin polymerization by mebendazole induces the loss of cytoplasmic microtubules, which leads to decreased glucose uptake and increased use of stored glycogen [23,24]. Genes related to glucose uptake in humans are known as the GLUT gene family, and it has been reported that the Caenorhabditis elegans-facilitated glucose transporter (FGT) gene in C. elegans performs functions similar to those of GLUT in humans [25,26]. However, there are no reports on GLUT or FGT-like genes in protozoa, including ciliates. In our study, the expression levels of GLUT1 and SLC2A3 (solute carrier family 2, facilitated glucose transporter member 3 genes) in humans and At5g16150 (the plastidic glucose transporter 4) gene in Arabidopsis thaliana were found to be decreased. However, the changes of expression levels of these genes were not statistically significant (p>0.05). The statistically non-significant downregulation in the expression of these genes is presumed to be related to the mode of action of metronidazole. The mode of action of benzimidazole derivatives involves the selective binding of helminths tubulin, thus inhibiting microtubule formation. Therefore, the effect of this drug may be slower than that of anthelmintics, which act as neurotransmitter agonists [27]. The expression levels of stress-related genes, such as catalase, were significantly decreased, and those of superoxide dismutase (Cu-Zn) genes, which serve antioxidants were also decreased but not significantly.
- Taken together, our results show that the intermittent use of mebendazole can be effective against M. avidus infection in vitro, although it can be different in salt waters. The gene expression profile after treatment with mebendazole revealed that most of the upregulated genes were related to membrane transport. The downregulated genes consisted of three main categories: tubulin, metabolism and transport-related groups. A couple of stress-related and glucose transporter-related genes were also downregulated but their expression was not statistically significant. These results suggest that the successful killing effect of mebendazole against M. avidus is due to changes in the membrane transporters and permeability. In addition, inhibition of tubulin polymerization and decreased metabolism have also been shown to play a role in its killing effect. Transcriptome analysis of mebendazole treatment against M. avidus will provide valuable genetic knowledge to explore the possibility of using mebendazole for scutica control.
Discussion
-
Conflicts of interest
Hee-Jae Cha is an editorial board member of the journal but was not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflicts of interest relevant to this article were reported.
-
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03933725)
This research was a part of the project titled “Omics based on fishery disease control technology development and industrialization (20150242),” funded by the Ministry of Oceans and Fisheries, Korea.
-
Author contributions
Conceptualization: HK. Data curation: ARL. Formal analysis: EJK. Funding acquisition: HJC, MSO. Methodology: HK, ARL, MSO. Project administration: MSO. Visualization: KYJ, EJK. Writing - original draft: ARL, MSO. Writing - review & editing: EJK, HJC, MSO. Approval of final manuscript: all authors.
Article information

| Category | Name | Description | log2 (FC) |
|---|---|---|---|
| Glucose transporter | At5g16150 | Plastidic glucose transporter 4 | –0.32 |
| TBA_TETTH | Glucose transporter type 1 | –2.08 | |
| SLC2A3 | Solute carrier family 2, facilitated glucose transporter member 3 | –2.29 | |
| Immune response-related | SOD1 | Superoxide dismutase (Cu-Zn) | –3.10 |
| Sod-1 | Superoxide dismutase (Cu-Zn) | –3.16 | |
| CSD2 | Superoxide dismutase (Cu-Zn) 2, chloroplastic | –10.00 | |
| kat | Catalase | –2.73 | |
| CAT2a) | Catalase-2 | –5.35 | |
| CAT3a) | Catalase-3 | –12.60 |
- 1. Jee BY, Shin KW, Lee DW, Kim YJ, Lee MK. Monitoring of the mortalities and medications in the inland farms of olive flounder, Paralichthys olivaceus, in South Korea. J Fish Pathol 2014;27:77–83.Article
- 2. Kang BJ, Jang YH, Jhon BK, Park BH, Jin CN. Monitoring of scuticociliatosis of olive Flounder (Paralichthys olivaceus) farm in Jeju, Korea from 2007 to 2014. J Fish Pathol 2015;28:165–9.Article
- 3. Retallack H, Okihiro MS, Britton E, Sommeran SV, DeRisi JL. Metagenomic next-generation sequencing reveals Miamiensis avidus (Ciliophora: Scuticociliatida) in the 2017 epizootic of leopard sharks (Triakis semifasciata) in San Francisco Bay, California, USA. J Wildl Dis 2019;55:375–86.ArticlePubMed
- 4. Jung SJ, Kitamura S, Song JY, Oh MJ. Miamiensis avidus (Ciliophora: Scuticociliatida) causes systemic infection of olive flounder Paralichthys olivaceus and is a senior synonym of Philasterides dicentrarchi. Dis Aquat Organ 2007;73:227–34.ArticlePubMed
- 5. Aquaculture Research Department, Aquatic Disease Control Division. Aquatic medicine catalog. Busan: National Institute of Fisheries Science (NIFS); 2016. p. 211.
- 6. Mariante RM, Vancini RG, Melo AL, Benchimol M. Giardia lamblia: evaluation of the in vitro effects of nocodazole and colchicine on trophozoites. Exp Parasitol 2005;110:62–72.ArticlePubMed
- 7. van der Werff SD, Vereecken K, van der Laan K, Campos Ponce M, Junco Diaz R, Nunez FA, et al. Impact of periodic selective mebendazole treatment on soil-transmitted helminth infections in Cuban schoolchildren. Trop Med Int Health 2014;19:706–18.ArticlePubMed
- 8. Chai JY, Jung BK, Hong SJ. Albendazole and mebendazole as anti-parasitic and anti-cancer agents: an update. Korean J Parasitol 2021;59:189–225.ArticlePubMedPMC
- 9. Locatelli C, Pedrosa RC, De Bem AF, Creczynski-Pasa TB, Cordova CA, Wilhelm-Filho D. A comparative study of albendazole and mebendazole-induced, time-dependent oxidative stress. Redox Rep 2004;9:89–95.ArticlePubMed
- 10. Kim KH, Park SI, Jee BY. Efficacy of oral administration of praziquantel and mebendazole against Microcotyle sebastis (Monogenea) infestation of cultured rockfish (Sebastes schlegeli). Fish Pathol 1998;33:467–71.Article
- 11. Chagas EC, de Araujo LD, Martins ML, Gomes LC, de Oliveira Malta JC, Varella AB, et al. Mebendazole dietary supplementation controls Monogenoidea (Platyhelminthes: Dactylogyridae) and does not alter the physiology of the freshwater fish Colossoma macropomum (Cuvier, 1818). Aquaculture 2016;464:185–9.Article
- 12. Whang I, Kang HS, Lee J. Identification of scuticociliates (Pseudocohnilembus persalinus, P. longisetus, Uronema marinum and Miamiensis avidus) based on the cox1 sequence. Parasitol Int 2013;62:7–13.ArticlePubMed
- 13. Iglesias R, Parama A, Alvarez MF, Leiro J, Sanmartin ML. Antiprotozoals effective in vitro against the scuticociliate fish pathogen Philasterides dicentrarchi. Dis Aquat Organ 2002;49:191–7.ArticlePubMed
- 14. Barrowman MM, Marriner SE, Bogan JA. The binding and subsequent inhibition of tubulin polymerization in Ascaris suum (in vitro) by benzimidazole anthelmintics. Biochem Pharmacol 1984;33:3037–40.ArticlePubMed
- 15. McKellar QA, Scott EW. The benzimidazole anthelmintic agents: a review. J Vet Pharmacol Ther 1990;13:223–47.ArticlePubMed
- 16. Andersson CR, Selvin T, Blom K, Rubin J, Berglund M, Jarvius M, et al. Mebendazole is unique among tubulin-active drugs in activating the MEK-ERK pathway. Sci Rep 2020;10:13124.ArticlePubMedPMC
- 17. Cowan AT, Bowman GR, Edwards KF, Emerson JJ, Turkewitz AP. Genetic, genomic, and functional analysis of the granule lattice proteins in Tetrahymena secretory granules. Mol Biol Cell 2005;16:4046–60.ArticlePubMedPMC
- 18. Agrawal A, Bisharyan Y, Papoyan A, Bednenko J, Cardarelli J, Yao M, et al. Fusion to Tetrahymena thermophila granule lattice protein 1 confers solubility to sexual stage malaria antigens in Escherichia coli. Protein Expr Purif 2019;153:7–17.ArticlePubMedPMC
- 19. Joffe LS, Schneider R, Lopes W, Azevedo R, Staats CC, Kmetzsch L, et al. The anti-helminthic compound mebendazole has multiple antifungal effects against Cryptococcus neoformans. Front Microbiol 2017;8:535.ArticlePubMedPMC
- 20. Schmahl G, Benini J. Treatment of fish parasites. 11. Effects of different benzimidazole derivatives (albendazole, mebendazole, fenbendazole) on Glugea anomala, Moniez, 1887 (Microsporidia): ultrastructural aspects and efficacy studies. Parasitol Res 1998;84:41–9.ArticlePubMedPDF
- 21. Thornalley PJ. Glyoxalase I: structure, function and a critical role in the enzymatic defence against glycation. Biochem Soc Trans 2003;31(Pt 6):1343–8.ArticlePubMedPDF
- 22. Reck-Peterson SL, Redwine WB, Vale RD, Carter AP. The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol 2018;19:382–98.ArticlePubMedPMC
- 23. Van den Bossche H, De Nollin S. Effects of mebendazole on the absorption of low molecular weight nutrients by Ascaris suum. Int J Parasitol 1973;3:401–7.ArticlePubMed
- 24. Lacey E. Mode of action of benzimidazoles. Parasitol Today 1990;6:112–5.ArticlePubMed
- 25. Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res 2002;35:9–26.ArticlePubMed
- 26. Kitaoka S, Morielli AD, Zhao FQ. FGT-1 is a mammalian GLUT2-like facilitative glucose transporter in Caenorhabditis elegans whose malfunction induces fat accumulation in intestinal cells. PLoS One 2013;8:e68475.ArticlePubMedPMC
- 27. Martin RJ. Modes of action of anthelmintic drugs. Vet J 1997;154:11–34.ArticlePubMed
References
Figure & Data
References
Citations


 KOSIN UNIVERSITY COLLEGE OF MEDICINE
KOSIN UNIVERSITY COLLEGE OF MEDICINE
 PubReader
PubReader ePub Link
ePub Link Cite
Cite